Sequencing Primer Design Tool Eurofins
Choosing suitable primers is one of the most important factors affecting 3 basic steps of the polymerase chain reaction process (PCR).
Specific amplification of the DNA template requires that designed primers do not have matches to other targets that allow undesired amplification. Primer design tools makes the design much more easy.
Primer3 design tool
Primer3 is the open-source software used for primer design. It is incorporated into various software packages and web services.
Primer3 can perform several kinds of design tasks and check correctness of existing primer pairs.
Primer3 chooses primers based on:
- melting temperature (Tm), GC content, primer-dimer possibilities;
- size of PCR product;
- positional constraints within DNA template, and so on.
Primer3 can also design hybridization probes and sequencing primers.
The components of Primer3 are the two web interfaces, Primer3web (the original web interface) and Primer3Plus (an easier-to-use, task-oriented interface), and the command-line primer-design program, primer3_core.
Primer3_core can be integrated with other software and useful to bioinformaticians.
The most recent primer3_core release is available at sourceforge site.
References:
- Untergasser A, Cutcutache I, Koressaar T, et al. Primer3—new capabilities and interfaces. Nucleic Acids Research. 2012;40(15):e115. doi:10.1093/nar/gks596.
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Research. 2007;35(Web Server issue):W71-W74. doi:10.1093/nar/gkm306.
- Primer3's documentation
NCBI/ Primer-BLAST design tool
Primer-BLAST is a web service that supports the selection of specific primers by considering possibilities for non-specific binding of primers across an entire genome or transcriptome.
It was developed at The National Center for Biotechnology Information (NCBI).
Primer-BLAST design tool uses Primer3 to design PCR primers and then uses BLAST (Basic Local Alignment Search Tool) and global alignment algorithm to sift them against user-selected database to avoid primer pairs that can cause non-specific amplifications.
Primer-BLAST allows users to design target-specific primers in one step and to check the specificity of existing primers.
References:
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi:10.1186/1471-2105-13-134.
- BLAST Homepage and Selected Search Pages
FastPCR primer design tool
FastPCR online is software tool for PCR primers design. It is written in Java and requires the Java 8 Runtime Environment (JRE) on a computer.
It cab be used for designing primers for most PCR applications - standard, multiplex, long-distance, inverse, real-time, group-specific, unique, and overlap extension PCR.
The primer design software analyzes primers for primer secondary structures such as G/C-quadruplexes (like G-G, G-T, G-A), hairpins, self-dimers and cross-dimers in primer pairs.
References:
- FastPCR Manual
- FastPCR on Wikipedia
- Kalendar R, Khassenov B, Ramankulov Y, Samuilova O, Ivanov KI. FastPCR: An in silico tool for fast primer and probe design and advanced sequence analysis. Genomics. 2017; 109(3-4):312-319. [pubmed]
IDT SciTools: PrimerQuest, OligoAnalyzer
PrimerQuest
PrimerQuest is the PCR primer and probe design tool of Integrated DNA Technologies, Inc., one of the leaders in oligonucleotide synthesis.
The PrimerQuest incorporates Primer3 software. It takes an input sequence in FASTA format and searches that sequence for optimal primers.
The PrimerQuest design tool allows to set reaction conditions, specify design regions, and so on.
OligoAnalyzer
OligoAnalyzer gives physical properties of a given oligo sequence.
The first screen allows entering the primer sequence which then can be analyzed for a possible presence of secondary structures and screened against selected databases with BLAST.
References:
- The Polymerase Chain Reaction by Integrated DNA Technologies, Inc.
GenScript molecular biology tools for PCR
GenScript Online PCR Primers Designs Tool
GenScript Online PCR Primers Designs Tooltakes an input sequence and allows to set reaction conditions depending on chosen option (basic, standard, advanced).
GenScript Real-time PCR (TaqMan) Primer Design
GenScript Real-time PCR (TaqMan) Primer Design serves to design primers and probes for Real-time PCR (TaqMan) experiments.
Adjustable options include:
- the PCR amplicon's size;
- Tm (melting temperature);
- the organism.
GenScript DNA Sequencing Primers Design Tool
GenScript DNA Sequencing Primers Design Tool helps to design primers for sequencing and allows to choose the approximate distance between sequencing primers and the Tm (melting temperature).
Eurofins Genomics LLC primers designs tools
Eurofins Genomics' primers designs tools is based on Prime+ of the GCG Wisconsin Package.
Eurofins Genomics' PCR Primer Design Tool
Eurofins Genomics' PCR Primer Design Tool has pre-set default values that can be changed to customize design parameters to the specific experiment.
Customizable design parameters include:
- Design Parameters (target region, primer 3′ clamp, max. Tm difference between forward and reverse primers)
- Primer Characteristics (length, GC content, Tm)
- Amplicon Characteristics (Size, GC content, Tm)
- Reaction Conditions
Eurofins Genomics' Sequencing Primer Design Tool
Eurofins Genomics' Sequencing Primer Design Tool allows to design forward and reverse sequencing primers for a target of interest.
Default conditions correspond to the optimal parameters for a primer and can be changed for a more tailored primer design.
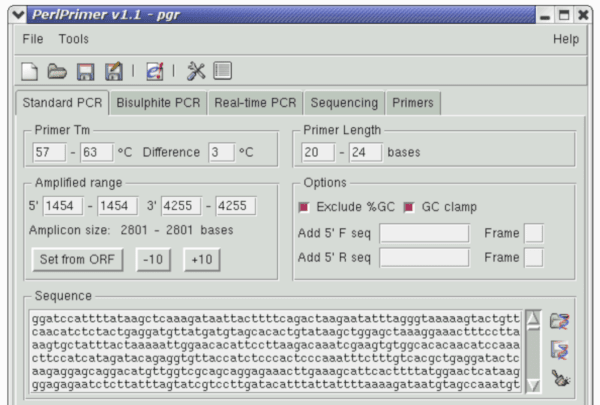
PerlPrimer design tool
PerlPrimer is the open-source primer design tool. It can be installed on both Microsoft Windows and Linux systems.
PerlPrimer is used for standard PCR, bisulphite PCR, real-time PCR (QPCR) and sequencing.
PerlPrimer uses melting-temperature and primer-dimer prediction algorithms as well as sequences retrieval from Ensembl and the ability to BLAST search primer pairs.
References:
- PerlPrimer manual
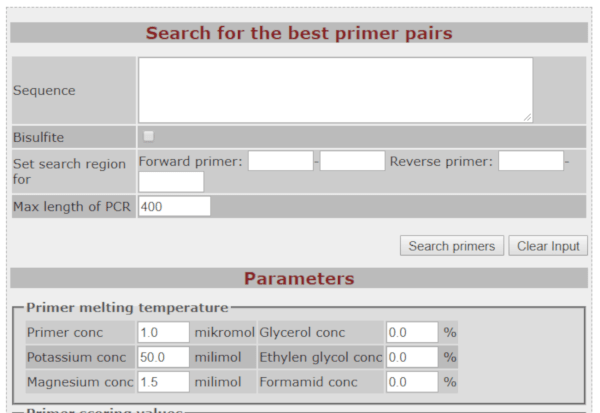
BiSearch Primer Design and Search Tool
BiSearch is a tool for primer-design of any DNA template. Furthermore, BiSearch primer tool can be useful to design primers for bisulfite-treated DNA templates, required to determine genomic DNA methylation profiles. It is composed of a primer-design and an electronic PCR (ePCR) algorithm.
References:
- BiSearch manual
Related articles
Sequencing Primer Design Tool Eurofins
Source: https://biology.reachingfordreams.com/biology/molecular-genetics/methods-in-molecular-genetics/84-dna-primer-design-tools
Posted by: hatcherdeconsenry.blogspot.com

0 Response to "Sequencing Primer Design Tool Eurofins"
Post a Comment